Abstract
INTRODUCTION: Multiple myeloma (MM) is preceded by an asymptomatic expansion of clonal plasma cells leading to precursor conditions labeled as monoclonal gammopathy of undetermined significance (MGUS) or smoldering multiple myeloma (SMM). The MM life history is characterized by a complex and heterogeneous evolution. Historically, hyperdiploidy (i.e. multiple trisomies on odd-numbered chromosomes) and translocations affecting immunoglobulin heavy chain (IGH) are recognized as the earliest and initiating genomic events, often acquired decades prior to diagnosis of MM. However, the potential existence of genomic events acquired before these known MM initiating events have never been investigated.
METHODS: Here, we isolated malignant plasma cells from bone marrow (BM) samples using CD138+ magnetic-bead or flowcytometry (CD38, CD138, and CD45) sorting from 58 newly diagnosed MM patients. All samples were interrogated by whole-genome sequencing (WGS) to investigate the existence of genomic events acquired before the IGH-translocations and hyperdiploidy. The cohort median coverage was coverage 75x (range 66-94).
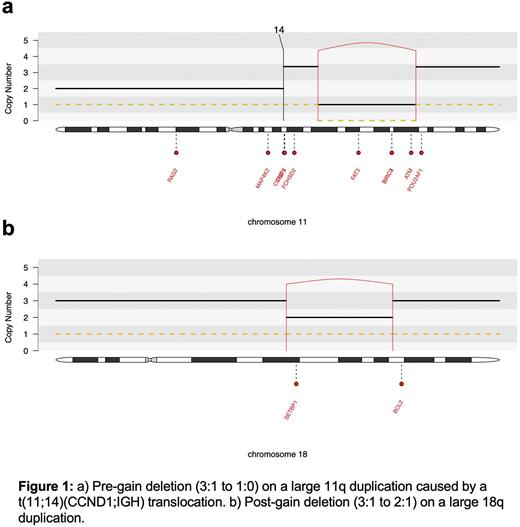
RESULTS: To determine whether we could detect potential driver events preceding early and initiating MM drivers, we developed a new chronological analytical model based on the integration of single nucleotide variants (SNV), structural variants (SV) and copy number variants (CNV). This model is composed by two main steps. First, we estimated the molecular time (i.e. corrected ratio between duplicated and non-duplicated clonal SNV) of all large chromosomal duplications and identified the earliest set of chromosomal gains in each patient life. In line with previous evidence (Maura et al. Nat Comm 2019), large trisomies were acquired in the early phases of MM evolution [median molecular time of the earliest gains 0.44 (range 0.10 - 0.75)]. Second, within each early large chromosomal gain we looked for clonal SV mediating CNV loss. A deletion on a large gain can generate three possible scenarios: 1) one of the two duplicated alleles is lost after the gain (i.e. post-gain event) causing a CNV jump from 3:1 to 2:1, where the first number represents the total alleles and the second the minor allele; 2) the deletion occurs on the minor and non-duplicated allele, causing a CNV jump from 3:1 to 2:0. In this scenario it is impossible to time the deletion in relation with the chromosomal duplication; 3) the duplicated allele is involved by a deletion before the duplication (i.e., pre-gain) causing a CNV jump from 3:1 to 1:0. When running this workflow, 5.2% (3/58) of patients in our cohort had large pre-gain deletions mediated by SVs acquired before the earliest multi-chromosomal gain events. All these events involved oncogenes such as ATM, BIRC3, USP8, TCF3. Post-gain loss mediated by SVs were observed in 48.3% (28/58) of patients. Surprisingly, in one patient we observed a pre-gain deletion (3:1 to 1:0) on a large chromosome 11q duplication caused by a t(11;14)(CCND1;IGH). Because the gain occurred simultaneously with the translocation, the deletion must have been acquired before both events. Overall, this is the first evidence that both hyperdiploid and t(11;14)(CCND1;IGH) can be preceded by earlier driver events.
To validate these findings, we performed the same analysis on the MMRF CoMMpass study (n=752 NDMM patients). Despite the low coverage of CoMMpass WGS not allowing for molecular time estimation, we identified pre-gain events and post-gain mediated by SVs in 10.1% (77/752) and 34.6% (260/752), respectively.
CONCLUSION: Due to the development of a new WGS-based chronological model, we revealed the existence of SV acquired before canonical "MM-initiating" events in newly diagnosed MM patients. This data suggests that there may be alternative, previously undescribed, MM-initiating events, which can be explored in a lager dataset using our novel analytical methods.
Disclosures
Diamond:Janssen: Honoraria; Medscape: Honoraria. Dogan:Incyte: Consultancy; Seattle Genetics: Consultancy; EUSA Pharma: Consultancy; Peer View: Honoraria; Takeda: Other: Research Funding; Roche: Other: Research Funding; Loxo: Consultancy; Physicians' Education Resource: Consultancy, Honoraria. Lesokhin:Janssen, Pfizer, Iteos, Sanofi, Genmab: Honoraria; Janssen, Pfizer, BMS, Genentech/Roche: Research Funding; BMS: Honoraria; Amgen: Honoraria; Serametrix, inc: Patents & Royalties; Sanofi: Research Funding; Trillium Therapeutics: Consultancy, Research Funding; Pfizer, Genmab, Sanofi, Iteos, BMS, Janssen: Consultancy; Memorial Sloan Kettering Cancer Center: Current Employment. Davies:Takeda, Abbvie, Amgen, BMS/Celgene, Sanofi, GSK, Janssen: Membership on an entity's Board of Directors or advisory committees. Korde:Amgen, Janssen: Research Funding; Clinical Care Options, OncLive, Intellisphere: Consultancy. Raab:Takeda: Membership on an entity's Board of Directors or advisory committees; Sanofi: Membership on an entity's Board of Directors or advisory committees; Novartis: Membership on an entity's Board of Directors or advisory committees; Heidelberg Pharma: Research Funding; BMS: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees. Usmani:Abbvie, Amgen, BMS, Celgene, EdoPharma, Genentech, Gilead, GSK, Janssen,Oncopeptides, Sanofi, Seattle Genetics, SecuraBio, SkylineDX, Takeda, TeneoBio: Consultancy; Amgen, BMS, Janssen, Sanofi: Speakers Bureau; Amgen, Array Biopharma, BMS, Celgene, GSK, Janssen, Merck, Pharmacyclics, Sanofi, Seattle Genetics, SkylineDX, Takeda: Research Funding. Landgren:Aptitude Health: Honoraria; Janssen: Honoraria, Other: Independent Data Monitoring Committee (IDMC) member for clinical trials, Research Funding; Rising Tide Foundation: Research Funding; Pfizer Inc: Consultancy; Leukemia & Lymphoma Society: Research Funding; MMRF: Honoraria; Theradex: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; NCI/NIH: Research Funding; Riney Foundation: Research Funding; Tow Foundation: Research Funding; Merck & Co., Inc.: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; Amgen: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal